Projects
- 1
Project A: | Complex-valued auto-encoders for self supervised quantitative MRI |
|
| Leader: | Chris Parker | CIG (CMIC) |
Deep-learning based quantitative MRI promises fast and reliable inference of tissue properties from MRI data. However, approaches so far have utilised the magnitude of complex MRI data as input for network training. This ignores phase information, which relates to biophysically relevant tissue properties, such as asymmetric diffusion and net flow. We will test the capability of complex-valued neural networks to encode such properties directly from the complex MRI data. As well as providing new tools for self-supervised quantitative MRI, the project will impart a deeper understanding of the diffusion-weighted MRI signal and how it relates the acquisition and underlying biophysical processes.
Project B: | Unlocking BBB Insights with GANs and autoencoders: Correcting Ktrans Values in DCE MRI for Enhanced Understanding of Inflammation-Related Brain Disorders |
|
| Leader: | Mara Cercignani | CUBRIC (Cardiff) |
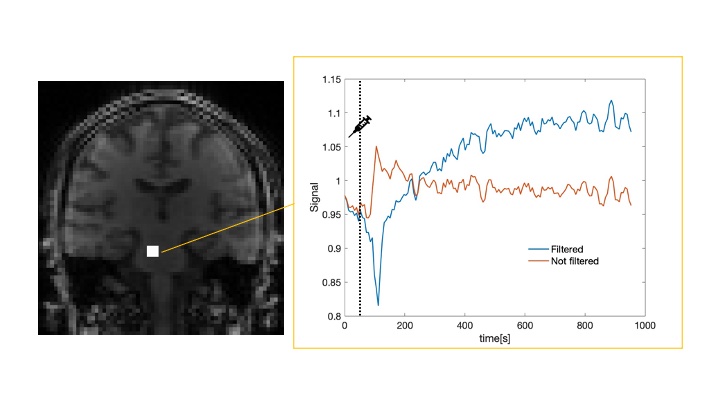
Dynamic Contrast-Enhanced Magnetic Resonance Imaging (DCE MRI) is a technique used to assess the permeability of the blood-brain barrier (BBB). The parameter of interest, known as Ktrans, is typically estimated by fitting a mathematical model to the data acquired from serial T1-weighted images taken after the gadolinium injection. In our study we have identified negative Ktrans values primarily attributed to filtering procedures applied by the MRI scanner. This project's core objective is to rectify these issues by learning the mapping between filtered and unfiltered data. We possess both filtered and unfiltered MRI data from the same acquisitions, which serves as the basis for our approach. To tackle this challenge, we would like to explore multiple avenues, including the possibility of training autoencoders or utilizing Generative Adversarial Networks (GANs) to map between filtered and unfiltered images. This project is expected to appeal to experts in Python and PyTorch, or equivalent deep learning libraries.
Project C: | Expanding a Multi-Modal Data Fusion Library with Fusilli: Adding Models and More |
|
| Leader: | Florence Townend | MANIFOLD (CMIC) |
We are currently developing a Python library called Fusilli, which is a collection of methods for multi-modal data fusion. Data fusion is the process of merging various data sources to achieve common goals. For example, using clinical data and MRI scans together to predict disease progression. The main use-case for Fusilli is for users to compare the performances of many fusion methods against each other for a prediction task. The library already offers around 15 data fusion methods, and we aim to add more during this hackathon. We aim to expand Fusilli by adding new models from and inspired by existing papers. If we have time, we will also be doing some unit-testing on the models and adding documentation. This project will give you hands-on experience in building Python libraries, from adding to the source code to unit testing to documentation. If you're not keen on adding methods, you can contribute by developing documentation like example notebooks and tutorials or by test-running the library on any appropriate data that you have already. Some familiarity with the python libraries PyTorch (Fusilli is written with PyTorch and PyTorch Lightning) and/or TensorFlow is a plus but not necessary.
Project D: | Using Network Spreading Models to Uncover Mechanisms of Alzheimer’s Disease Progression |
|
| Leaders: | Ellie Thompson, Anna Schroder, Tiantian He and Neil Oxtoby | POND (CMIC) |
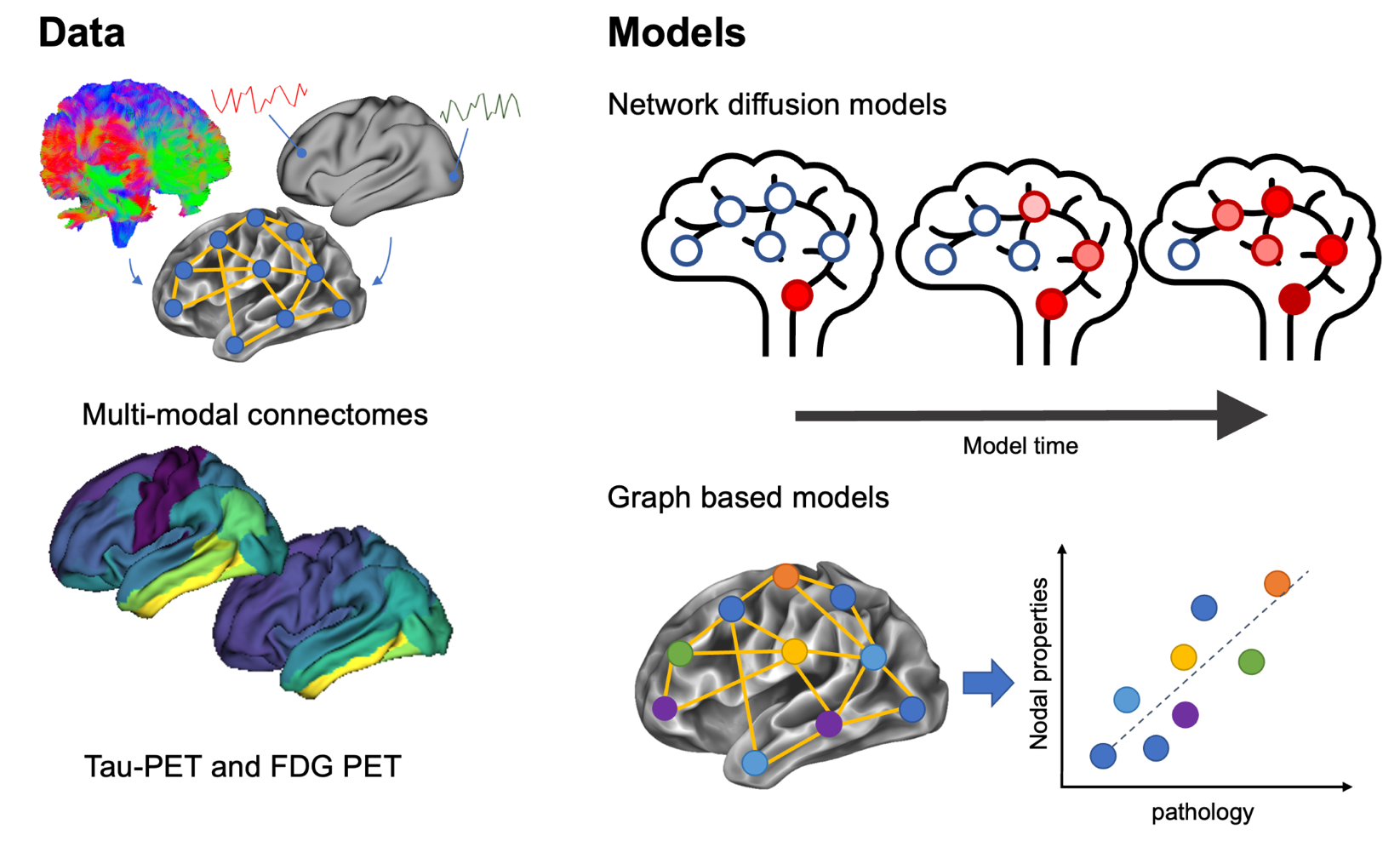
Connectome-based models are used to probe mechanisms of pathology spread in neurodegenerative disease, and to predict patient-specific outcomes (see here for a recent review). Recent work using these models indicates early stages of Alzheimer’s disease are dominated by spreading, whereas later stages are dominated by production of tau (see here). models have been proposed, but there is no framework available to easily compare them. This project aims to create a toolbox containing different models to allow the user to compare model fits with different models and parameters. In addition, we plan to use the framework to see whether abnormal glucose metabolism (measured by FDG PET) mediates pathology accumulation (measured by tau PET), e.g., spreading vs production.
Full project details, including useful skills and learning outcomes can be found here.
Project E: | Hacking Real-time AI workflows for Surgery |
|
| Leaders: | Miguel Xochicale and Zhehua Mao | COMPASS, Surgical Robot Vision (WEISS) and ARC |
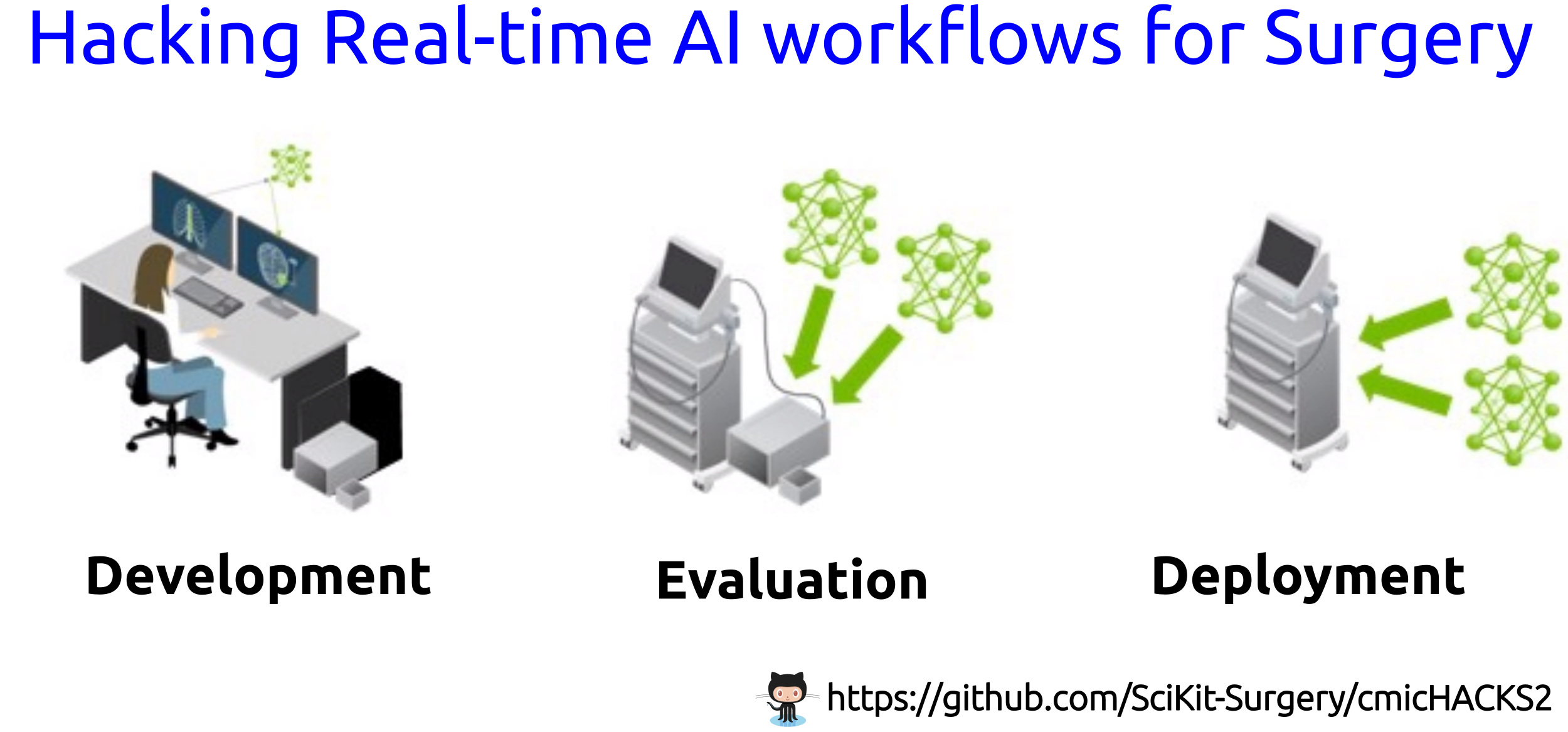
Artificial Intelligence (AI)-based surgical workflows use input from multiple sources of data (e.g., medical devices, trackers, robots, cameras, etc) with different Machine Learning and Deep Learning tasks (e.g., classification, segmentation, synthesis, etc) and have been applied across different surgical and acquisition workflows. However, the diversity of data sources, pre-processing methods, training and inference methods make a challenging scenario for low-latency applications in surgery. In this hackathon, instructors aim to engage with participants to bring three learning outcomes: (a) participants will learn to train, optimise, test and deploy AI models for detection and tool tracking, (b) participants will learn good software practices to contribute to our open-source projects aligning with medical device software standard (IEC 62304), and (c) participants will have the chance to work with NVIDIA Clara AGX - A Universal computing architecture for next-generation AI medical instruments. We hope to bring together researchers, engineers and clinicians across different departments to hack workflows for real-time AI for Surgery (development, evaluation and integration) and hopefully to spark future collaborations. For further information such as requirements, resources, discussion forum and more please visit https://github.com/SciKit-Surgery/cmicHACKS2.
Project F: | GPU-enabled Monte Carlo simulations of flow and diffusion |
|
| Leaders: | Lizzie Powell and Leevi Kerkela | QIG (CMIC) and GOSH |
Background: There are several Monte Carlo simulators for modelling spin diffusion in tissue microstructure and the resulting MR signals; Camino is a popular example, developed here at UCL. However, all current simulation toolboxes ignore a key feature of tissue microstructure - the vasculature! Lizzie has recently been working to address this issue, designing a toolbox that can model both diffusion and flow spin dynamics inside a novel microvascular substrate mesh. However, this proof-of-concept design has been implemented in Matlab, which is notoriously slow...
Aim: To integrate the same flow modelling architecture into disimpy, a massively parallelised, open source, GPU-enabled Python toolbox for diffusion simulations (paper, github) developed by Leevi. We will use novel microvascular meshes to define the direction of flow, combine flow and diffusive spin dynamics, and enable realistic IVIM-style simulations in reasonable computation times.
Anticipated outputs: Conference abstract, CDMRI paper
Useful skills: Python & CUDA
Project G: | Latent space representations of disease progression |
|
| Leaders: | Alex Young, Andre Altmann and Liza Levitis | POND / COMBINE (CMIC) |
Disease progression modelling constructs latent timelines from disease biomarker data that can be used to identify an individual’s disease stage and other factors that influence their biomarker profile (such as disease subtypes). Across disciplines, many other techniques similarly construct a latent space that can be used to explore different sources of variability. Examples include trajectory inference methods used in single cell omics, variational auto encoders, principal components analysis, and normative models. The aim of this project is to stimulate new ideas for how we can adapt these techniques for disease progression modelling and other tasks. To do this, the project brings together researchers from across CMIC to compare the similarities and differences between different latent space representations output by a variety of techniques. We will apply as many techniques as possible to a common dataset and then compare their outputs and performance for different tasks. For this project we are looking to build a team across a diverse range of expertise and a variety of programming languages.
Project H: | Self-Supervised Superheroes - To ssVERDICT and Beyond! |
|
| Leader: | Snigdha Sen | Ccami (CMIC) |
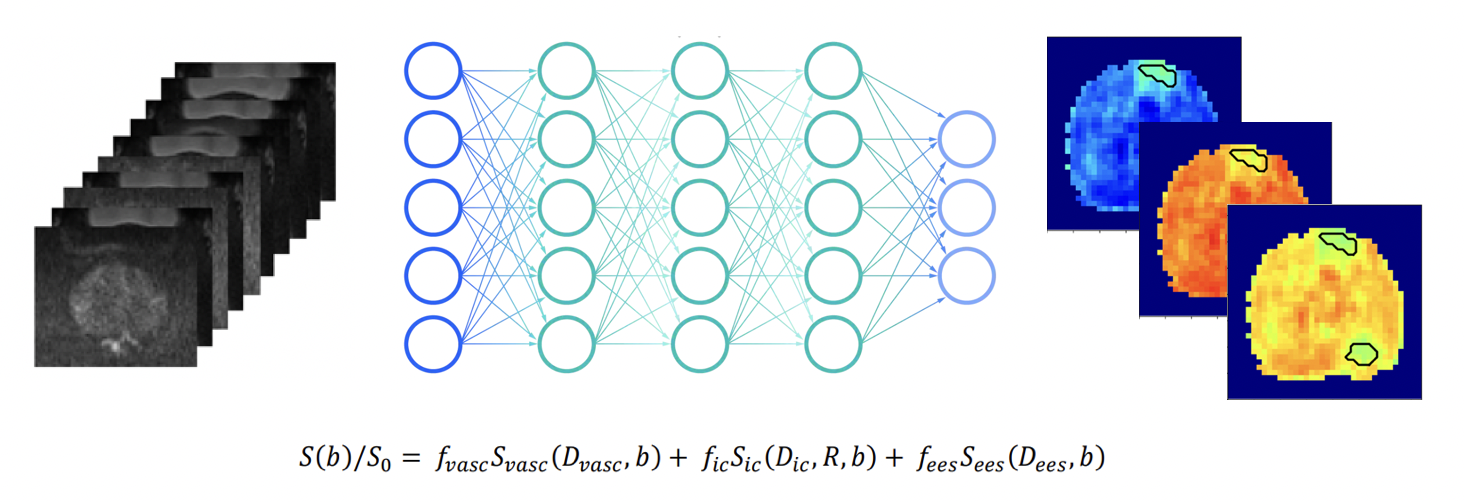
Computational models of the diffusion MRI signal can be fitted to the data to estimate the models’ parameter values in the tissue, and we can compare the values in healthy tissue vs disease to improve non-invasive diagnosis. There has been a move towards fitting these models using deep learning, but supervised methods can be significantly biased by the training data. Self-supervised learning has been used successfully to improve estimation with the VERDICT model (three compartment model for prostate), see ssVERDICT arxiv. Currently, this work is coded up just to fit the VERDICT model. In this project, we would like to expand and adapt the ssVERDICT code base to incorporate a wide range of diffusion MRI compartment models, which can be combined to form complex biophysical models (e.g. NODDI, SANDI and more) as required. If time allows, we will also investigate using different network architectures for the fitting (e.g. VAE, CNN) and make the code flexible to different fitting methods. The end result would be a flexible software package for fitting any diffusion MRI model with self-supervised learning with a variety of network architectures. It would be useful to have experience using Python, PyTorch and GitHub.
Project I: | Abnormality Detection in OCT Scans through Normative Modeling on Latent Diffusion Autoencoder |
|
| Leader: | Mariam Zabihi | MANIFOLD (CMIC) |
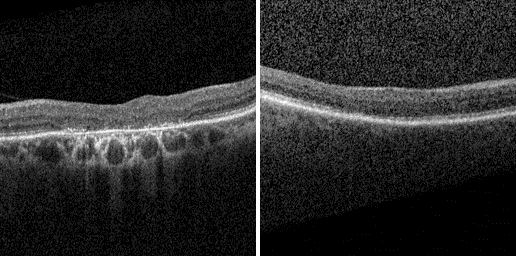
Research Question:
Can the application of normative modeling on the latent space of a Diffusion Autoencoder significantly enhance abnormality detection in OCT scans?
Objective:
To develop and apply a normative model on the latent space of a Diffusion Autoencoder (DAE) for advanced detection of abnormalities, with a specific focus on OCT scans. This project aims to demonstrate how this integration can lead to more accurate and reliable identification of retinal conditions like HCQ retinopathy.
Method:
Diffusion Autoencoder: optimize a Diffusion Autoencoder tailored for precise reconstruction of OCT images and extraction of intricate latent representations.
Normative Modeling on Latent Space: Implement a normative model within the latent space of the DAE. This model will establish a statistical baseline of what constitutes a "normal" or healthy OCT scan.
Abnormality Detection Mechanism: Develop a system that leverages the normative model to identify deviations from the norm, signaling potential abnormalities in OCT scans.
Dataset Preparation: Gather and preprocess a dataset of OCT scans, ensuring a balance of healthy and pathological cases for training and validation.
Model Training and Evaluation: Train the system using the prepared dataset, and rigorously evaluate its performance in detecting abnormalities, particularly focusing on HCQ retinopathy.
Expected Outcome: A sophisticated abnormality detection system that combines the generative capabilities of a Diffusion Autoencoder with the precision of normative modeling.
Program
Download
Location
The hackathon is a purely in-person event which will take place in:
Jeffery (Thursday) and Elvin (Friday) rooms,
Level 1, main UCL Institute of Education (IOE) building,
20 Bedford Way, London WC1H 0AL.
On Thursday, at the end of day, participants are invited to socialize with food and drinks in:
The Marquis Cornwallis,
31 Marchmont St, Greater, London WC1N 1AP.